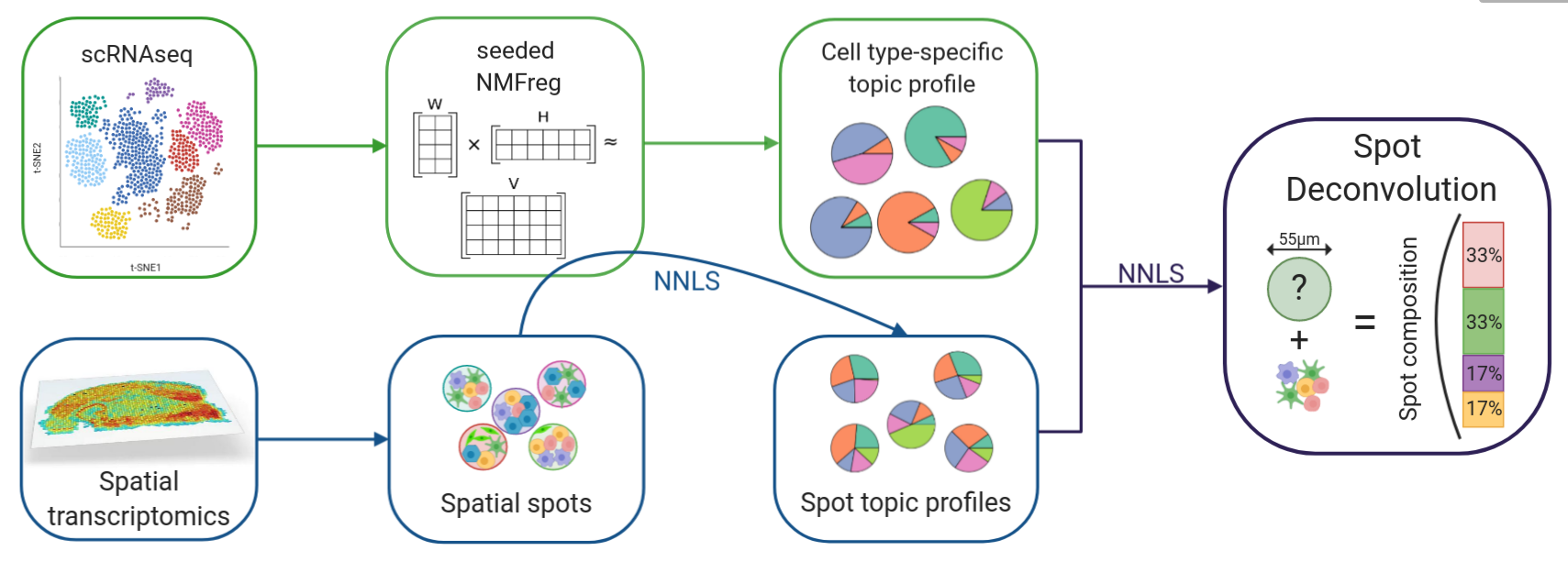
SPOTlight: Deconvolution of VISIUM data from singlecell reference
Installation
Prerequisite
scGPT works with Python >= 3.7.13 and R >=3.6.1. Please make sure you have the correct version of Python and R installed pre-installation.
conda create -n scGPT_py39 python=3.9 conda activate scGPT_py39
conda create -n scGPT python=3.8
conda activate scGPT
conda search r-base
conda install -c conda-forge r-base=4.3.3
PyPI
1
pip3 install scgpt "flash-attn<1.0.5" "orbax<0.1.8"
pip3 install packaging pip3 install torch conda install cuda -c nvidia # ml cuda, ml cudnn pip3 install llvmlite==0.38.0
pip3 install scgpt pip3 install wandb
pip3 install “flash-attn<1.0.5”
cuda 11.7
conda install nvidia/label/cuda-11.7.0::cuda-toolkit
bsub -P acc_scMultiscale -q interactive -n 1 -R v100 -gpu num=1 -W 06:00 -Is /bin/bash
conda create -n scGPT python=3.10.11 conda activate scGPT ml proxies ml cuda/11.7.0 #nvcc –version
https://developer.nvidia.com/cuda-11-7-0-download-archive
gpt서버 아니어도 설치됨
conda install cudatoolkit=11.7 cudatoolkit-dev ‘gxx>=6.0.0,<12.0’ cudnn pip3 install torch==1.13.0 pip3 install torchvision==0.14.0 pip3 install torchaudio==0.13.0
#python -m pip install setuptools==69.5.1 pip install packaging conda install conda-forge::git
#pip install ninja pip install “flash-attn<1.0.5” –no-build-isolation conda install r-base r-devtools pip install –no-deps scgpt pip install ipykernel python -m ipykernel install –user –name=scGPT pip install pandas pip install scanpy pip install scvi-tools==0.16.0 #pip install numba –upgrade pip install numpy==1.24.4 pip install torchtext==0.14.0 pip install scib pip install datasets pip install wandb pip install pyro-ppl==1.8.2 pip install cell-gears==0.0.1 pip install orbax==0.1.7
Python 3.10.11 | packaged by conda-forge | (main, May 10 2023, 18:58:44) [GCC 11.3.0] on linux Type “help”, “copyright”, “credits” or “license” for more information.
import copy import gc import json import os from pathlib import Path import shutil import sys import time import traceback from typing import List, Tuple, Dict, Union, Optional import warnings import pandas as pd import pickle import torch from anndata import AnnData import scanpy as sc import scvi Global seed set to 0 import seaborn as sns import numpy as np import wandb from scipy.sparse import issparse import matplotlib.pyplot as plt from torch import nn from torch.nn import functional as F from torch.utils.data import Dataset, DataLoader from sklearn.model_selection import train_test_split from sklearn.metrics import adjusted_rand_score, normalized_mutual_info_score from torchtext.vocab import Vocab from torchtext._torchtext import ( … Vocab as VocabPybind, … ) from sklearn.metrics import confusion_matrix
sys.path.insert(0, “../”) import scgpt as scg from scgpt.model import TransformerModel, AdversarialDiscriminator from scgpt.tokenizer import tokenize_and_pad_batch, random_mask_value from scgpt.loss import ( … masked_mse_loss, … masked_relative_error, … criterion_neg_log_bernoulli, … ) from scgpt.tokenizer.gene_tokenizer import GeneVocab from scgpt.preprocess import Preprocessor from scgpt import SubsetsBatchSampler from scgpt.utils import set_seed, category_str2int, eval_scib_metrics
sc.set_figure_params(figsize=(6, 6)) os.environ[“KMP_WARNINGS”] = “off” warnings.filterwarnings(‘ignore’) hyperparameter_defaults = dict( … seed=0, … dataset_name=”ms”, … do_train=True, … load_model=”./save/scGPT_human”, … mask_ratio=0.0, … epochs=10, … n_bins=51, … MVC=False, # Masked value prediction for cell embedding … ecs_thres=0.0, # Elastic cell similarity objective, 0.0 to 1.0, 0.0 to disable … dab_weight=0.0, … lr=1e-4, … batch_size=32, … layer_size=128, … nlayers=4, # number of nn.TransformerEncoderLayer in nn.TransformerEncoder … nhead=4, # number of heads in nn.MultiheadAttention … dropout=0.2, # dropout probability … schedule_ratio=0.9, # ratio of epochs for learning rate schedule … save_eval_interval=5, … fast_transformer=True, … pre_norm=False, … amp=True, # Automatic Mixed Precision … include_zero_gene = False, … freeze = False, #freeze … DSBN = False, # Domain-spec batchnorm … )
run = wandb.init( … config=hyperparameter_defaults, … project=”scGPT”, … reinit=True, … settings=wandb.Settings(start_method=”fork”), … ) wandb: (1) Create a W&B account wandb: (2) Use an existing W&B account wandb: (3) Don’t visualize my results wandb: Enter your choice: 1 wandb: You chose ‘Create a W&B account’ wandb: Create an account here: https://wandb.ai/authorize?signup=true wandb: Paste an API key from your profile and hit enter, or press ctrl+c to quit: wandb: Appending key for api.wandb.ai to your netrc file: /hpc/users/chos14/.netrc wandb: Tracking run with wandb version 0.17.1 wandb: Run data is saved locally in /sc/arion/projects/scMultiscale/chos14/scGPT/wandb/run-20240616_205706-8i6qrhaw wandb: Run
wandb offlineto turn off syncing. wandb: Syncing run flowing-wind-1 wandb: ⭐️ View project at https://wandb.ai/icahnmssm/scGPT wandb: 🚀 View run at https://wandb.ai/icahnmssm/scGPT/runs/8i6qrhaw config = wandb.config print(config) {‘seed’: 0, ‘dataset_name’: ‘ms’, ‘do_train’: True, ‘load_model’: ‘../save/scGPT_human’, ‘mask_ratio’: 0.0, ‘epochs’: 10, ‘n_bins’: 51, ‘MVC’: False, ‘ecs_thres’: 0.0, ‘dab_weight’: 0.0, ‘lr’: 0.0001, ‘batch_size’: 32, ‘layer_size’: 128, ‘nlayers’: 4, ‘nhead’: 4, ‘dropout’: 0.2, ‘schedule_ratio’: 0.9, ‘save_eval_interval’: 5, ‘fast_transformer’: True, ‘pre_norm’: False, ‘amp’: True, ‘include_zero_gene’: False, ‘freeze’: False, ‘DSBN’: False}set_seed(config.seed)
settings for input and preprocessing
pad_token = “
include_zero_gene = config.include_zero_gene # if True, include zero genes among hvgs in the training max_seq_len = 3001 n_bins = config.n_bins
input/output representation
input_style = “binned” # “normed_raw”, “log1p”, or “binned” output_style = “binned” # “normed_raw”, “log1p”, or “binned”
settings for training
MLM = False # whether to use masked language modeling, currently it is always on. CLS = True # celltype classification objective ADV = False # Adversarial training for batch correction CCE = False # Contrastive cell embedding objective MVC = config.MVC # Masked value prediction for cell embedding ECS = config.ecs_thres > 0 # Elastic cell similarity objective DAB = False # Domain adaptation by reverse backpropagation, set to 2 for separate optimizer INPUT_BATCH_LABELS = False # TODO: have these help MLM and MVC, while not to classifier input_emb_style = “continuous” # “category” or “continuous” or “scaling” cell_emb_style = “cls” # “avg-pool” or “w-pool” or “cls” adv_E_delay_epochs = 0 # delay adversarial training on encoder for a few epochs adv_D_delay_epochs = 0 mvc_decoder_style = “inner product” ecs_threshold = config.ecs_thres dab_weight = config.dab_weight
explicit_zero_prob = MLM and include_zero_gene # whether explicit bernoulli for zeros do_sample_in_train = False and explicit_zero_prob # sample the bernoulli in training
per_seq_batch_sample = False
settings for optimizer
lr = config.lr # TODO: test learning rate ratio between two tasks lr_ADV = 1e-3 # learning rate for discriminator, used when ADV is True batch_size = config.batch_size eval_batch_size = config.batch_size epochs = config.epochs schedule_interval = 1
settings for the model
fast_transformer = config.fast_transformer fast_transformer_backend = “flash” # “linear” or “flash” embsize = config.layer_size # embedding dimension d_hid = config.layer_size # dimension of the feedforward network in TransformerEncoder nlayers = config.nlayers # number of TransformerEncoderLayer in TransformerEncoder nhead = config.nhead # number of heads in nn.MultiheadAttention dropout = config.dropout # dropout probability
logging
log_interval = 100 # iterations save_eval_interval = config.save_eval_interval # epochs do_eval_scib_metrics = True
%% validate settings
assert input_style in [“normed_raw”, “log1p”, “binned”]
assert output_style in [“normed_raw”, “log1p”, “binned”]
assert input_emb_style in [“category”, “continuous”, “scaling”]
if input_style == “binned”:
if input_emb_style == “scaling”:
raise ValueError(“input_emb_style scaling is not supported for binned input.”)
elif input_style == “log1p” or input_style == “normed_raw”:
if input_emb_style == “category”:
raise ValueError(
“input_emb_style category is not supported for log1p or normed_raw input.”
)
if input_emb_style == “category”: mask_value = n_bins + 1 pad_value = n_bins # for padding gene expr values n_input_bins = n_bins + 2 else: mask_value = -1 pad_value = -2 n_input_bins = n_bins
if ADV and DAB: raise ValueError(“ADV and DAB cannot be both True.”)
DAB_separate_optim = True if DAB > 1 else False
dataset_name = config.dataset_name save_dir = Path(f”./save/dev_{dataset_name}-{time.strftime(‘%b%d-%H-%M’)}/”) save_dir.mkdir(parents=True, exist_ok=True) print(f”save to {save_dir}”) logger = scg.logger scg.utils.add_file_handler(logger, save_dir / “run.log”)
Step 2: Load and pre-process data
if dataset_name == “ms”:
data_dir = Path(“data/ms”)
adata = sc.read(data_dir / “c_data.h5ad”)
adata_test = sc.read(data_dir / “filtered_ms_adata.h5ad”)
adata.obs[“celltype”] = adata.obs[“Factor Value[inferred cell type - authors labels]”].astype(“category”)
adata_test.obs[“celltype”] = adata_test.obs[“Factor Value[inferred cell type - authors labels]”].astype(“category”)
adata.obs[“batch_id”] = adata.obs[“str_batch”] = “0”
adata_test.obs[“batch_id”] = adata_test.obs[“str_batch”] = “1”
adata.var.set_index(adata.var[“gene_name”], inplace=True)
adata_test.var.set_index(adata.var[“gene_name”], inplace=True)
data_is_raw = False
filter_gene_by_counts = False
adata_test_raw = adata_test.copy()
adata = adata.concatenate(adata_test, batch_key=”str_batch”)
make the batch category column
batch_id_labels = adata.obs[“str_batch”].astype(“category”).cat.codes.values adata.obs[“batch_id”] = batch_id_labels celltype_id_labels = adata.obs[“celltype”].astype(“category”).cat.codes.values celltypes = adata.obs[“celltype”].unique() num_types = len(np.unique(celltype_id_labels)) id2type = dict(enumerate(adata.obs[“celltype”].astype(“category”).cat.categories)) adata.obs[“celltype_id”] = celltype_id_labels adata.var[“gene_name”] = adata.var.index.tolist()
if config.load_model is not None: model_dir = Path(config.load_model) model_config_file = model_dir / “args.json” model_file = model_dir / “best_model.pt” vocab_file = model_dir / “vocab.json”
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
vocab = GeneVocab.from_file(vocab_file)
shutil.copy(vocab_file, save_dir / "vocab.json")
for s in special_tokens:
if s not in vocab:
vocab.append_token(s)
adata.var["id_in_vocab"] = [
1 if gene in vocab else -1 for gene in adata.var["gene_name"]
]
gene_ids_in_vocab = np.array(adata.var["id_in_vocab"])
logger.info(
f"match {np.sum(gene_ids_in_vocab >= 0)}/{len(gene_ids_in_vocab)} genes "
f"in vocabulary of size {len(vocab)}."
)
adata = adata[:, adata.var["id_in_vocab"] >= 0]
# model
with open(model_config_file, "r") as f:
model_configs = json.load(f)
logger.info(
f"Resume model from {model_file}, the model args will override the "
f"config {model_config_file}."
)
embsize = model_configs["embsize"]
nhead = model_configs["nheads"]
d_hid = model_configs["d_hid"]
nlayers = model_configs["nlayers"]
n_layers_cls = model_configs["n_layers_cls"]
PosixPath(‘save/dev_ms-Jun16-21-16/vocab.json’) scGPT - INFO - match 2808/2808 genes in vocabulary of size 60697. scGPT - INFO - Resume model from save/scGPT_human/best_model.pt, the model args will override the config save/scGPT_human/args.json.
set up the preprocessor, use the args to config the workflow
preprocessor = Preprocessor( use_key=”X”, # the key in adata.layers to use as raw data filter_gene_by_counts=filter_gene_by_counts, # step 1 filter_cell_by_counts=False, # step 2 normalize_total=1e4, # 3. whether to normalize the raw data and to what sum result_normed_key=”X_normed”, # the key in adata.layers to store the normalized data log1p=data_is_raw, # 4. whether to log1p the normalized data result_log1p_key=”X_log1p”, subset_hvg=False, # 5. whether to subset the raw data to highly variable genes hvg_flavor=”seurat_v3” if data_is_raw else “cell_ranger”, binning=n_bins, # 6. whether to bin the raw data and to what number of bins result_binned_key=”X_binned”, # the key in adata.layers to store the binned data )
adata_test = adata[adata.obs[“str_batch”] == “1”] adata = adata[adata.obs[“str_batch”] == “0”]
preprocessor(adata, batch_key=None)
scGPT - INFO - Normalizing total counts … scGPT - INFO - Binning data …
preprocessor(adata_test, batch_key=None)
scGPT - INFO - Normalizing total counts … scGPT - INFO - Binning data …
input_layer_key = { # the values of this map coorespond to the keys in preprocessing “normed_raw”: “X_normed”, “log1p”: “X_normed”, “binned”: “X_binned”, }[input_style] all_counts = ( adata.layers[input_layer_key].A if issparse(adata.layers[input_layer_key]) else adata.layers[input_layer_key] ) genes = adata.var[“gene_name”].tolist()
celltypes_labels = adata.obs[“celltype_id”].tolist() # make sure count from 0 celltypes_labels = np.array(celltypes_labels)
batch_ids = adata.obs[“batch_id”].tolist() num_batch_types = len(set(batch_ids)) batch_ids = np.array(batch_ids)
( train_data, valid_data, train_celltype_labels, valid_celltype_labels, train_batch_labels, valid_batch_labels, ) = train_test_split( all_counts, celltypes_labels, batch_ids, test_size=0.1, shuffle=True )
if config.load_model is None: vocab = Vocab( VocabPybind(genes + special_tokens, None) ) # bidirectional lookup [gene <-> int]
vocab.set_default_index(vocab[“
tokenized_train = tokenize_and_pad_batch(
train_data,
gene_ids,
max_len=max_seq_len,
vocab=vocab,
pad_token=pad_token,
pad_value=pad_value,
append_cls=True, # append
scGPT - INFO - train set number of samples: 7059, feature length: 1309
logger.info( f”valid set number of samples: {tokenized_valid[‘genes’].shape[0]}, “ f”\n\t feature length: {tokenized_valid[‘genes’].shape[1]}” )
scGPT - INFO - valid set number of samples: 785, feature length: 1338
def prepare_data(sort_seq_batch=False) -> Tuple[Dict[str, torch.Tensor]]: masked_values_train = random_mask_value( tokenized_train[“values”], mask_ratio=mask_ratio, mask_value=mask_value, pad_value=pad_value, ) masked_values_valid = random_mask_value( tokenized_valid[“values”], mask_ratio=mask_ratio, mask_value=mask_value, pad_value=pad_value, ) print( f”random masking at epoch {epoch:3d}, ratio of masked values in train: “, f”{(masked_values_train == mask_value).sum() / (masked_values_train - pad_value).count_nonzero():.4f}”, ) input_gene_ids_train, input_gene_ids_valid = ( tokenized_train[“genes”], tokenized_valid[“genes”], ) input_values_train, input_values_valid = masked_values_train, masked_values_valid target_values_train, target_values_valid = ( tokenized_train[“values”], tokenized_valid[“values”], ) tensor_batch_labels_train = torch.from_numpy(train_batch_labels).long() tensor_batch_labels_valid = torch.from_numpy(valid_batch_labels).long() tensor_celltype_labels_train = torch.from_numpy(train_celltype_labels).long() tensor_celltype_labels_valid = torch.from_numpy(valid_celltype_labels).long() if sort_seq_batch: # TODO: update to random pick seq source in each traning batch train_sort_ids = np.argsort(train_batch_labels) input_gene_ids_train = input_gene_ids_train[train_sort_ids] input_values_train = input_values_train[train_sort_ids] target_values_train = target_values_train[train_sort_ids] tensor_batch_labels_train = tensor_batch_labels_train[train_sort_ids] tensor_celltype_labels_train = tensor_celltype_labels_train[train_sort_ids] valid_sort_ids = np.argsort(valid_batch_labels) input_gene_ids_valid = input_gene_ids_valid[valid_sort_ids] input_values_valid = input_values_valid[valid_sort_ids] target_values_valid = target_values_valid[valid_sort_ids] tensor_batch_labels_valid = tensor_batch_labels_valid[valid_sort_ids] tensor_celltype_labels_valid = tensor_celltype_labels_valid[valid_sort_ids] train_data_pt = { “gene_ids”: input_gene_ids_train, “values”: input_values_train, “target_values”: target_values_train, “batch_labels”: tensor_batch_labels_train, “celltype_labels”: tensor_celltype_labels_train, } valid_data_pt = { “gene_ids”: input_gene_ids_valid, “values”: input_values_valid, “target_values”: target_values_valid, “batch_labels”: tensor_batch_labels_valid, “celltype_labels”: tensor_celltype_labels_valid, } return train_data_pt, valid_data_pt
dataset
class SeqDataset(Dataset): def init(self, data: Dict[str, torch.Tensor]): self.data = data def len(self): return self.data[“gene_ids”].shape[0] def getitem(self, idx): return {k: v[idx] for k, v in self.data.items()}
data_loader
def prepare_dataloader( data_pt: Dict[str, torch.Tensor], batch_size: int, shuffle: bool = False, intra_domain_shuffle: bool = False, drop_last: bool = False, num_workers: int = 0, ) -> DataLoader: if num_workers == 0: num_workers = min(len(os.sched_getaffinity(0)), batch_size // 2) dataset = SeqDataset(data_pt) if per_seq_batch_sample: # find the indices of samples in each seq batch subsets = [] batch_labels_array = data_pt[“batch_labels”].numpy() for batch_label in np.unique(batch_labels_array): batch_indices = np.where(batch_labels_array == batch_label)[0].tolist() subsets.append(batch_indices) data_loader = DataLoader( dataset=dataset, batch_sampler=SubsetsBatchSampler( subsets, batch_size, intra_subset_shuffle=intra_domain_shuffle, inter_subset_shuffle=shuffle, drop_last=drop_last, ), num_workers=num_workers, pin_memory=True, ) return data_loader data_loader = DataLoader( dataset=dataset, batch_size=batch_size, shuffle=shuffle, drop_last=drop_last, num_workers=num_workers, pin_memory=True, ) return data_loader
Step 3: Load the pre-trained scGPT model
device = torch.device(“cuda” if torch.cuda.is_available() else “cpu”)
ntokens = len(vocab) # size of vocabulary model = TransformerModel( ntokens, embsize, nhead, d_hid, nlayers, nlayers_cls=3, n_cls=num_types if CLS else 1, vocab=vocab, dropout=dropout, pad_token=pad_token, pad_value=pad_value, do_mvc=MVC, do_dab=DAB, use_batch_labels=INPUT_BATCH_LABELS, num_batch_labels=num_batch_types, domain_spec_batchnorm=config.DSBN, input_emb_style=input_emb_style, n_input_bins=n_input_bins, cell_emb_style=cell_emb_style, mvc_decoder_style=mvc_decoder_style, ecs_threshold=ecs_threshold, explicit_zero_prob=explicit_zero_prob, use_fast_transformer=fast_transformer, fast_transformer_backend=fast_transformer_backend, pre_norm=config.pre_norm, ) if config.load_model is not None: try: model.load_state_dict(torch.load(model_file)) logger.info(f”Loading all model params from {model_file}”) except: # only load params that are in the model and match the size model_dict = model.state_dict() pretrained_dict = torch.load(model_file) pretrained_dict = { k: v for k, v in pretrained_dict.items() if k in model_dict and v.shape == model_dict[k].shape } for k, v in pretrained_dict.items(): logger.info(f”Loading params {k} with shape {v.shape}”) model_dict.update(pretrained_dict) model.load_state_dict(model_dict)
pre_freeze_param_count = sum(dict((p.data_ptr(), p.numel()) for p in model.parameters() if p.requires_grad).values())
Freeze all pre-decoder weights
for name, para in model.named_parameters(): print(“-“*20) print(f”name: {name}”) if config.freeze and “encoder” in name and “transformer_encoder” not in name: # if config.freeze and “encoder” in name: print(f”freezing weights for: {name}”) para.requires_grad = False
post_freeze_param_count = sum(dict((p.data_ptr(), p.numel()) for p in model.parameters() if p.requires_grad).values())
logger.info(f”Total Pre freeze Params {(pre_freeze_param_count )}”) logger.info(f”Total Post freeze Params {(post_freeze_param_count )}”) wandb.log( { “info/pre_freeze_param_count”: pre_freeze_param_count, “info/post_freeze_param_count”: post_freeze_param_count, }, )
model.to(device) wandb.watch(model)
if ADV: discriminator = AdversarialDiscriminator( d_model=embsize, n_cls=num_batch_types, ).to(device)