[10X CellRanger] Custom Reference Genome: Multi Species
10X genomics에서 CellRanger 사용관련한 tutorial? support의 일환으로 사람(Homo sapiens, GRCh38)과 쥐(Mus musculus, mm10)에 대한 Reference를 제공하고 있다.1
하지만 그 외의 종이나 일반적인 genome 이외의 sequence를 align하고 분석하고 싶다면 Custom Reference를 만들어야 한다.
TdTomato sequence로 custom ref만들기 포스트
👉 여기 : 원래 있던 reference에 한두 서열 끼워넣어서 새로 만드는 방법
이번 포스트에서는 완전히 다른 종(개, canine)의 reference genome을 다운 받아서 만들어보려고 한다.
Input: Fasta & GTF files
Reference를 만들기 위해서는 (mkref의 input으로는) 만들려는 종의 fasta와 GTF file이 필요하다.
- Fasta (.fa)
- 그 종의 reference genome sequence를 chromosome 별로 정리해 놓은 파일. 항상
>이 기호로 시작해서 chr 이름이 첫줄에 들어가고, 다음줄엔 sequence가 저장돼 있는 format.cat xx.fa | grep '>'해서 chromosome이름을 확인할 수 있다. - GTF (.gtf)
- 유전자 정보가 저장된 파일. 각 유전자의 위치(chr:position), ensembl ID, coding/intron 등등의 정보가 저장돼 있다.
Canin .fa & .gtf
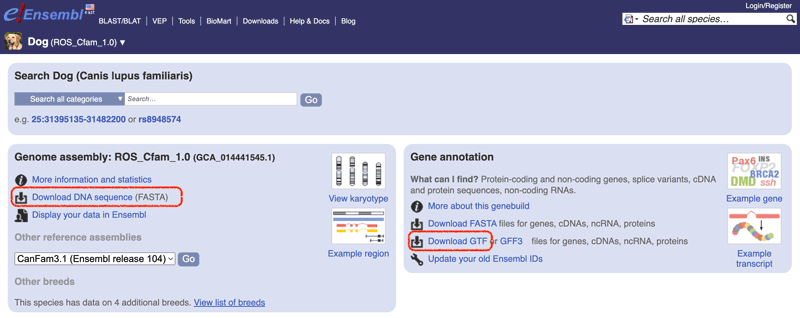
두 input file은 Ensembl database에서 쉽게 구할 수 있다.
1
2
3
4
5
6
7
8
## fasta file
wget https://ftp.ensembl.org/pub/release-109/fasta/canis_lupus_familiaris/dna/Canis_lupus_familiaris.ROS_Cfam_1.0.dna.toplevel.fa.gz
## gtf file
wget https://ftp.ensembl.org/pub/release-109/gtf/canis_lupus_familiaris/Canis_lupus_familiaris.ROS_Cfam_1.0.109.gtf.gz
## 압축해제
gunzip Canis_lupus_familiaris.ROS_Cfam_1.0.dna.toplevel.fa.gz
gunzip Canis_lupus_familiaris.ROS_Cfam_1.0.109.gtf.gz
Run mkref
👉 CellRanger mkref_Build Notes for Reference Packages
10X genomics download page에서 reference build tutorial을 약간 변형해서 진행
dog_fasta_in부분에 방금 다운받은 fasta file /dog_gtf_url부분에 gtf file 을 넣어준다.- 개 ENSEMBL gene은 ENGCAF로 시작하기 때문에
ID부분을 고쳐준다
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
#################### SETUP ####################
human_genome="GRCh38"
dog_genome="Canis10K"
version="2023-May"
build="GRCh38_and_Canis10K_2023-May_build"
mkdir -p "$build"
# Download source files if they do not exist in reference_sources/ folder
source="reference_sources"
mkdir -p "$source"
human_fasta_url="http://ftp.ensembl.org/pub/release-98/fasta/homo_sapiens/dna/Homo_sapiens.GRCh38.dna.primary_assembly.fa.gz"
human_fasta_in="${source}/Homo_sapiens.GRCh38.dna.primary_assembly.fa"
human_gtf_url="http://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_human/release_32/gencode.v32.primary_assembly.annotation.gtf.gz"
human_gtf_in="${source}/gencode.v32.primary_assembly.annotation.gtf"
#dog_fasta_url=""
dog_fasta_in="${source}/Canis_lupus_familiaris.ROS_Cfam_1.0.dna.toplevel.fa"
#dog_gtf_url=""
dog_gtf_in="${source}/Canis_lupus_familiaris.ROS_Cfam_1.0.109.gtf"
if [ ! -f "$human_fasta_in" ]; then
curl -sS "$human_fasta_url" | zcat > "$human_fasta_in"
fi
if [ ! -f "$human_gtf_in" ]; then
curl -sS "$human_gtf_url" | zcat > "$human_gtf_in"
fi
#if [ ! -f "$dog_fasta_in" ]; then
# curl -sS "$dog_fasta_url" | zcat > "$dog_fasta_in"
#fi
#if [ ! -f "$dog_gtf_in" ]; then
# curl -sS "$dog_gtf_url" | zcat > "$dog_gtf_in"
#fi
# String patterns used for both genomes
ID="(ENS(CAF)?[GTE][0-9]+)\.([0-9]+)"
BIOTYPE_PATTERN=\
"(protein_coding|lncRNA|\
IG_C_gene|IG_D_gene|IG_J_gene|IG_LV_gene|IG_V_gene|\
IG_V_pseudogene|IG_J_pseudogene|IG_C_pseudogene|\
TR_C_gene|TR_D_gene|TR_J_gene|TR_V_gene|\
TR_V_pseudogene|TR_J_pseudogene)"
GENE_PATTERN="gene_type \"${BIOTYPE_PATTERN}\""
GENE_PATTERN_DOG="gene_biotype \"${BIOTYPE_PATTERN}\""
TX_PATTERN="transcript_type \"${BIOTYPE_PATTERN}\""
TX_PATTERN_DOG="transcript_biotype \"${BIOTYPE_PATTERN}\""
READTHROUGH_PATTERN="tag \"readthrough_transcript\""
PAR_PATTERN="tag \"PAR\""
#################### HUMAN ####################
# Please see the GRCh38-2020-A build documentation for details on these steps.
# Process FASTA -- translate chromosome names
human_fasta_modified="$build/$(basename "$human_fasta_in").modified"
cat "$human_fasta_in" \
| sed -E 's/^>(\S+).*/>\1 \1/' \
| sed -E 's/^>([0-9]+|[XY]) />chr\1 /' \
| sed -E 's/^>MT />chrM /' \
> "$human_fasta_modified"
# Process GTF -- split Ensembl IDs from version suffixes
human_gtf_modified="$build/$(basename "$human_gtf_in").modified"
cat "$human_gtf_in" \
| sed -E 's/gene_id "'"$ID"'";/gene_id "\1"; gene_version "\3";/' \
| sed -E 's/transcript_id "'"$ID"'";/transcript_id "\1"; transcript_version "\3";/' \
| sed -E 's/exon_id "'"$ID"'";/exon_id "\1"; exon_version "\3";/' \
> "$human_gtf_modified"
# Process GTF -- filter based on gene/transcript tags
cat "$human_gtf_modified" \
| awk '$3 == "transcript"' \
| grep -E "$GENE_PATTERN" \
| grep -E "$TX_PATTERN" \
| grep -Ev "$READTHROUGH_PATTERN" \
| grep -Ev "$PAR_PATTERN" \
| sed -E 's/.*(gene_id "[^"]+").*/\1/' \
| sort \
| uniq \
> "${build}/gene_allowlist"
human_gtf_filtered="${build}/$(basename "$human_gtf_in").filtered"
grep -E "^#" "$human_gtf_modified" > "$human_gtf_filtered"
grep -Ff "${build}/gene_allowlist" "$human_gtf_modified" \
>> "$human_gtf_filtered"
#################### Dog ####################
# Process FASTA -- translate chromosome names
dog_fasta_modified="$build/$(basename "$dog_fasta_in").modified"
cat "$dog_fasta_in" \
| sed -E 's/^>(\S+).*/>\1 \1/' \
| sed -E 's/^>([0-9]+|[XY]) />\1 /' \
| sed -E 's/^>MT />M /' \
> "$dog_fasta_modified"
# Process GTF -- split Ensembl IDs from version suffixes
dog_gtf_modified="$build/$(basename "$dog_gtf_in").modified"
cat "$dog_gtf_in" \
| sed -E 's/gene_id "'"$ID"'";/gene_id "\1"; gene_version "\3";/' \
| sed -E 's/transcript_id "'"$ID"'";/transcript_id "\1"; transcript_version "\3";/' \
| sed -E 's/exon_id "'"$ID"'";/exon_id "\1"; exon_version "\3";/' \
> "$dog_gtf_modified"
# Process GTF -- filter based on gene/transcript tags
cat "$dog_gtf_modified" \
| awk '$3 == "transcript"' \
| grep -E "$GENE_PATTERN_DOG" \
| grep -E "$TX_PATTERN_DOG" \
| grep -Ev "$READTHROUGH_PATTERN" \
| sed -E 's/.*(gene_id "[^"]+").*/\1/' \
| sort \
| uniq \
> "${build}/gene_allowlist"
dog_gtf_filtered="${build}/$(basename "$dog_gtf_in").filtered"
grep -E "^#" "$dog_gtf_modified" > "$dog_gtf_filtered"
grep -Ff "${build}/gene_allowlist" "$dog_gtf_modified" \
>> "$dog_gtf_filtered"
#################### MKREF ####################
cellranger mkref --ref-version="$version" \
--genome="$human_genome" --fasta="$human_fasta_modified" --genes="$human_gtf_filtered" \
--genome="$dog_genome" --fasta="$dog_fasta_modified" --genes="$dog_gtf_filtered"
Result
- run time : 약 30 min
1
2
3
4
5
6
7
8
9
10
11
# result_dir 01
GRCh38_and_Canis10K_2023-May_build/
├── Canis_lupus_familiaris.ROS_Cfam_1.0.109.gtf.filtered
├── Canis_lupus_familiaris.ROS_Cfam_1.0.109.gtf.modified
├── Canis_lupus_familiaris.ROS_Cfam_1.0.dna.toplevel.fa.modified
├── gencode.v32.primary_assembly.annotation.gtf.filtered
├── gencode.v32.primary_assembly.annotation.gtf.modified
├── gene_allowlist
└── Homo_sapiens.GRCh38.dna.primary_assembly.fa.modified
0 directories, 7 files
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
# result_dir 02
GRCh38_and_Canis10K
├── fasta
│ ├── genome.fa
│ └── genome.fa.fai
├── genes
│ └── genes.gtf.gz
├── reference.json
└── star
├── chrLength.txt
├── chrNameLength.txt
├── chrName.txt
├── chrStart.txt
├── exonGeTrInfo.tab
├── exonInfo.tab
├── geneInfo.tab
├── Genome
├── genomeParameters.txt
├── SA
├── SAindex
├── sjdbInfo.txt
├── sjdbList.fromGTF.out.tab
├── sjdbList.out.tab
└── transcriptInfo.tab
3 directories, 19 files
이렇게 확인하는 code
1
tree -L 2 {directory_name}
Reference
This post is licensed under
CC BY 4.0
by the author.