SingleCell RNA-seq data로 GSEA하는법
FindMarker()로 DEG구한후에 어떤 function을 갖고 Gene set enrichment가 있는지 확인해보자
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
library(clusterProfiler)
library(AnnotationDbi)
library(org.Hs.eg.db)
library(fgsea)
library(presto)
library(tibble)
markers_young <- read.table("Mono.WilcoxDEG.final.txt")
markers_young_sig <- markers_young[markers_young$label%in%c("young_specific_up_deg","young_specific_down_deg"),]
markers_young_sig <- markers_young_sig[(markers_young_sig$avg_log2FC > 1) | (markers_young_sig$avg_log2FC < -0.4), ]
#up 493 / down 349
gene_list <- markers_young_sig$Young.avg_log2FC
names(gene_list) <- rownames(markers_young_sig)
gene_list <- sort(gene_list, decreasing = TRUE)
gse.Y <- gseGO(geneList = gene_list,
OrgDb = org.Hs.eg.db,
ont = "BP",
keyType = "SYMBOL", # Adjust the keyType based on your gene IDs
pvalueCutoff = 0.05,
verbose = TRUE,
pAdjustMethod = "BH",scoreType = "std",eps = 0)
result_df <- as.data.frame(gse.Y@result)
result_df <- result_df %>% dplyr::filter(setSize >= 5 & setSize <= 500)
# Select top 5 and bottom 5 based on NES
top_entries <- result_df %>% arrange(desc(NES)) %>% head(5)
bottom_entries <- result_df %>% arrange(desc(NES)) %>% tail(5)
top_bottom_entries <- rbind(top_entries, bottom_entries)
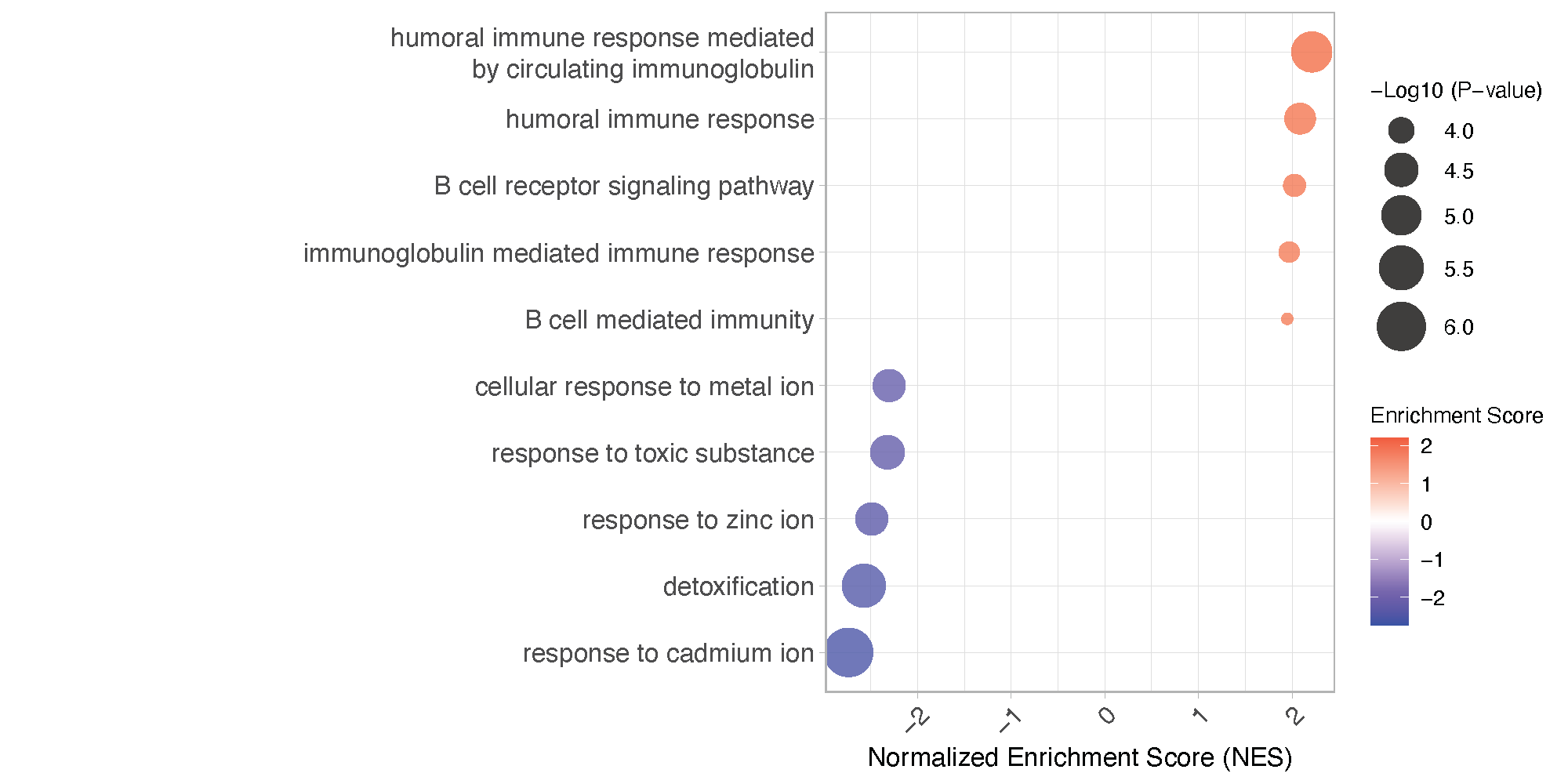
dot_plot <- ggplot(top_bottom_entries, aes(x = NES, y = reorder(Description, NES), size = -log10(pvalue), color = NES)) +
geom_point(alpha = 0.7) +
scale_size_continuous(name = "-Log10 (P-value)", range = c(2, 10)) +
scale_color_gradient2(low = "blue", mid = "white", high = "red", midpoint = 0, name = "Enrichment Score") +
labs(x = "Normalized Enrichment Score (NES)", y = "") + theme_light() +
theme(
text = element_text(size = 18), # General text size
axis.title = element_text(size = 12), # Axis titles
axis.text = element_text(size = 12), # Axis text
axis.text.x = element_text(angle = 45, hjust = 1), # Adjust x-axis text
legend.title = element_text(size = 10), # Legend title
legend.text = element_text(size = 10) # Legend text
)
ggsave(filename = "Wilcox.Young.GSEA.dot.pdf", plot = dot_plot, width = 10, height = 5, dpi = 300)
This post is licensed under
CC BY 4.0
by the author.